BIDSconvertR
 | The hexagonal sticker was made using the iconspackage and based on the MRI svg graphics provided by Flaticon and mavadee FlaticonLink. |
Table of contents
Aim
BIDSconvertR aims to provide a workflow that can:
- do the task inside of the R environment
- convert DICOM data to NIfTI (with the awesome dcm2niix (v1.0.20230411) or install a custom version)
- structure this data according to the BIDS specification
- validate the compatibility with BIDS with the BIDS validator
- visualize the images with papayaViewer and papayaWidget
Features
- continuous application
- lazy processing of already existing files
- easy application during data collection in ongoing studies
- user-friendliness (minimal terminal interaction required)
- user dialog with message boxes guiding the users through the workflow
- Shiny App (GUI) for sequence editing and data inspection
- file cleaning
- Renaming of subject-IDs or session-IDs with regular expressions.
- Renaming of session-IDs
- BIDS validation
- verification (color-coded) of sequence-IDs (comparing the entered sequence-IDs to regular expressions according to BIDS)
- implemented validation with BIDS-Validator (Website/Docker)
- quality control
- user-friendly (papayaWidget image viewer) for BIDS datasets
- pseudonymized BIDS output (only metadata)
- all potentially identifiable metadata was removed from images and JSONs
Only the metadata contained within the BIDS folder is free of potentially identifiable information. Follow the legal terms when sharing your dataset and consider additional defacing, pseudonymization, or both.
Workflow visualization
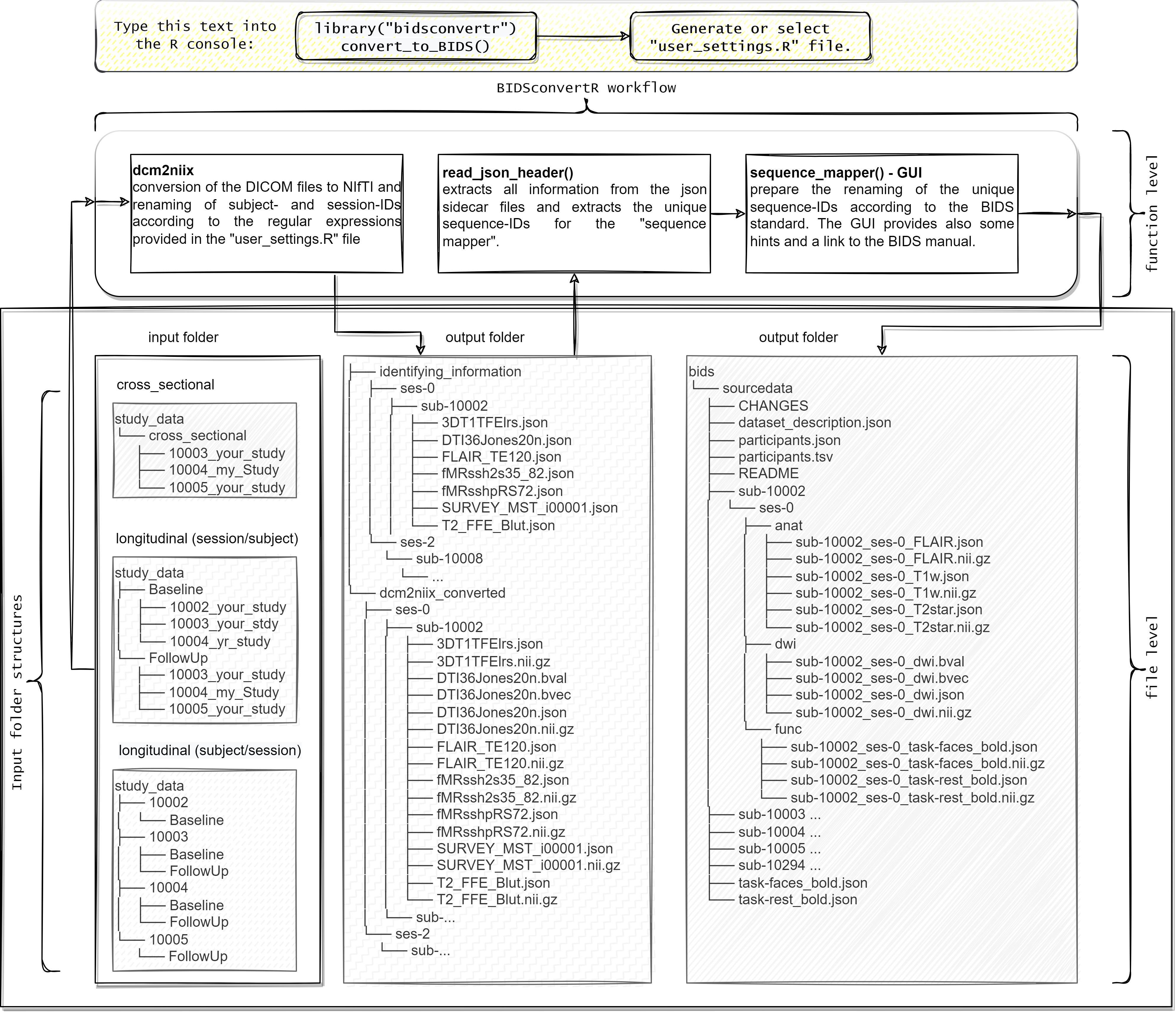
How to cite:
Wulms N, Eppe S, Dehghan‑Nayyeri M, Streeter A, Bonberg N, Berger K, Sundermann B, Minnerup H. (2023). The R package for DICOM to brain imaging data structure conversion Nature Scientific Data. 2023 Oct 04. doi: 10.1038/s41597‑023‑02583‑4 https://doi.org/10.1038/s41597-023-02583-4
Wulms, Niklas. (2023). BIDSconvertR. Zenodo. https://doi.org/10.5281/ZENODO.5878407
Milestones / To-Do’s
- create Docker container
- move image viewer into separate package
- adapt code to CRAN requirements


